Our Research
The Zhou Lab aims to achieve an in-depth understanding of regulatory mechanisms of epitranscriptome that can lead to discoveries of novel drug targets and diversify the toolbox of gene therapy.
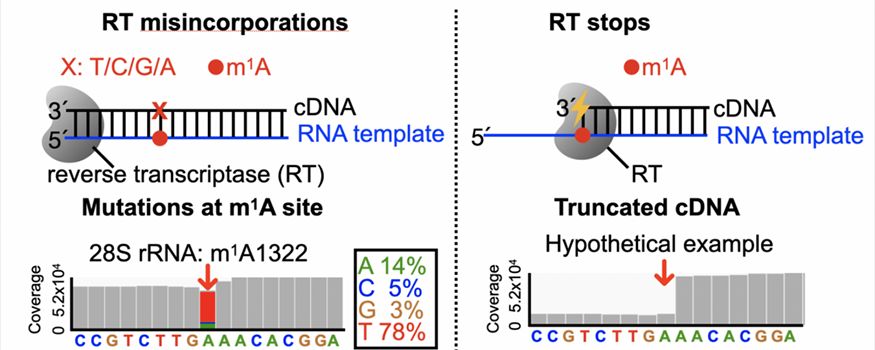
RT signatures (i.e., stops or misincorporations) allow detection of chemical modifications via sequencing.
RNA chemical modifications remain challenging to be detected precisely and quantitatively due to their diverse chemical nature, and the highly transient and dynamic nature of their carrier – endogenous RNA. Recent advances in the high-throughput sequencing technologies enable the rapid development of new methods that allow detection of RNA chemical modifications at base-resolution within the entire transcriptome.
The principle of the detection of chemical modifications by RNA-seq-based technologies relies on a unique and robust sequencing signature that a certain type of chemical modification can convert into. Most frequently, this signature is generated by reverse transcriptase. For chemical modifications that block canonical Waston-Crick-Frankin base-pairs, such as the N1-methyladenosine (m1A), they would naturally give rise to either stop or mutation signatures during reverse transcription (RT). For modifications that do not block canonical base-pairing such as N6-methyladenosine (m6A), one can apply chemical or enzymatic approaches to treat such modification in order to introduce secondary modifications that can generate RT signatures specifically at the modified rather than unmodified RNA.
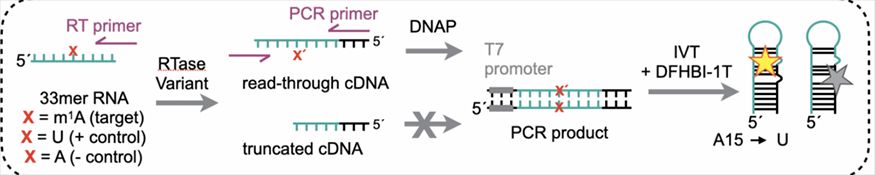
Directed Evolution Platform for engineering reverse transcriptases that can misincorporate at RNA modifications (Zhou et al Nat. Methods 2019).
We take approaches of a recently developed directed evolution platform to engineer reverse transcriptase that can directly encode any RNA modification into RT mutation signatures, in combination with next-generation sequencing technologies, to enable transcriptome-wide mapping of chemical modifications in RNA at base resolution. Such an effort will help eliminate the necessity of introducing secondary modification groups with additional chemical or enzymatic treatment and enhance the detection sensitivity and reproducibility. With quantitative measurements of modification stoichiometry, we are able to detect changes of the chemically modified transcripts in different cellular stages, such as during stem cell differentiation and cellular stress response, to reveal interplays between the modification level and the expression level of effector proteins, and to further understand the biological functions of modifications in fundamental biological processes.
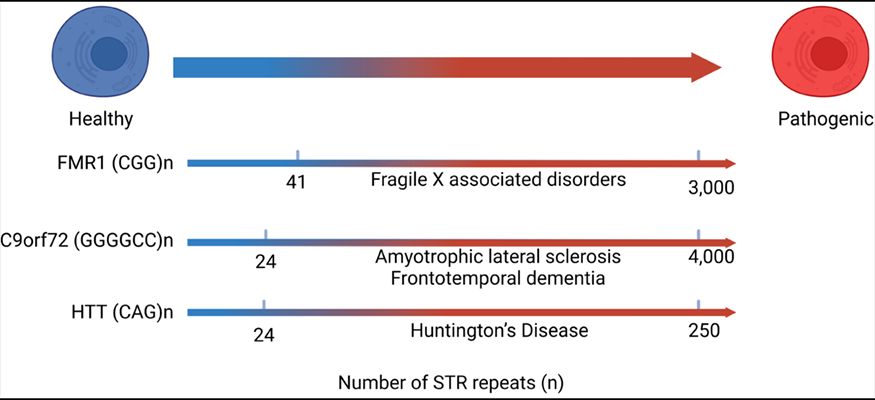
Short tandem repeats (STR) represent characteristic sequences consisting of a repeated pattern of 1-6 nucleotides, occupying around 3% of the human genome. Mutations such as repeat expansion of STR sequences often cause dysregulations in cellular functions and lead to diseases, particularly neurological disorders. STR RNA is indeed the central player in two primary pathological mechanisms proposed with many STR-associated conditions. One is called the RNA gain-of-function (GOF) mechanism. Mutated STR RNAs are bound with binding proteins to form inclusion bodies leading to aggregated RNA foci, a hallmark of many neurological disorders. The other one is through repeats-associated non-AUG translation (RAN), in which STR RNAs can be translated to hydrophobic peptides such as polyglycine that can trigger aggregation. Despite the central roles of STR RNAs, investigations of functions of STR RNAs in pathogenesis remain largely hampered by poorly characterized STR RNAs in disease-associated contexts regarding both primary sequences and secondary structures. STR RNAs form highly stable secondary structures that halt the reverse transcriptase (RTases), making them hard to amplify and detect. Therefore, in mechanistic studies for the STR-associated diseases, one must assume the RNA STR contains the same repeat number as that in the DNA template rather than directly determine the STR sequence. This project aims to develop an engineering platform to develop functional reverse transcriptase that allows robust read-through over stable RNA secondary structures focusing on those formed by STR RNA with a disease-associated number of repeats and allow accurate and sensitive detection of STR RNA.